Tutorial
Submitting a job
To use MINT WebServer you need to specify a PDB code with RNA or DNA structure or upload a PDB file containing your molecule.
In the first case, the .pdb file will be automatically downloaded from the PDB database and protonated using Reduce software. In the latter case, make sure that your structure contains hydrogens.
Filling the fields with an email address and a job title is necessary to access your results in the future.
Accessing results
After submitting a job you will be redirected to the Results page. You will see a table containing outputs of current and previous calculations.
The table may be accessed later, by clicking the Results tab and providing an email that was used during submission. However, bear in mind that we can not guarantee long-term data storage.
Outputs
Each row in the results table represents one job. The description of the columns is the following:
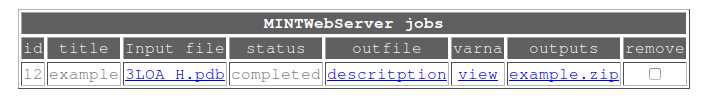

- Id is an internal MINT WebServer numbering of submitted jobs.
- title is the job name given during submission.
- input file gives the name of the analyzed structure. The structure may be an uploaded file or automatically protonated structure downloaded from the PDB database.
- status of the job may be queued, running, completed or canceled. The latter means that the provided structure could not be analyzed. Such an event is automatically reported to MINT authors.
- outfile is a raw text file named description containing all data calculated in the run.
- varna contains a link view to 2D visualizations of the computed parameters.
- outputs gives a link to the zipped package containing all files generated by MINT during the analysis.
- remove box allows selecting the results to be deleted from the database.
Output files
MINT WebServer produces multiple output files.
In all of them nucleotides are described in a manner that allows to identify them unambiguously.
We use the representation containing a chain name, a residue name and a residue number.
For example: the N|GUA:521 represents residuum number 521 from the chain N, with the name GUA.
The values posted in the chain name|residue name:residue number code are derived from the input PDB file.
The zipped package contains the following files:
_description - the main file containing the complete description of the structure. The file content starts from the list of all of the used parameters, followed by the list of helices, motifs, triplets and pseudo-knots. One can also find the list of both Watson-Crick (WC-WC) and non-Watson-Crick interactions, with the exact parameters of the hydrogen bonds. Additionally there is a simple dot-bracket representation of the secondary structure that can be used for visualization or energy computation by other tools ( _description example ).
_helices.csv - a file that contains a list of the deceted helices ( _helices.csv example).
_ion_pi.csv - a file that contains a list of the recognized ion-pi interactions (i.e., detected if a phosphorus atom in one nucleotide is placed close to the nucleobase of the second nucleotide) with their interaction energy values ( _ion_pi.csv example).
_motifs.csv - a file that contains list of the all RNA structural motifs ( _motifs.csv example)
_nucleotides_eval.csv - a file that contains the number of the WC-WC hydrogen bonds, non-WC-WC hydrogen bonds, Coulomb energy, van der Waals energy and their sum per nucleotide. The content gives the energetic description of the state of every nucleotide ( _nucleotides_eval.csv example).
_pairs.csv - a file that contains list of all detected pairs along with its classification according to edge (WC, Hoosteen, Sugar) and configuration (Cis or Trans) ( _pairs.csv example).
_pseudo.csv - a file that contains list of the all detected pseudo-knot ( _pseudo.csv example).
_stacking.csv - a file that contains a list of all recognized stacking pairs with their interaction energy values ( _stacking.csv example).
_triplets.csv - a file that contains the number of the recognized triplets ( _triplets.csv example).
_MINT.xls - a multiple sheets file that combines the above .csv files ( _MINT.xls example).
.pdb files - the analyzed structure with the column containing the beta-factors replaced with: the number of WC-WC hydrogen bonds (_2D.pdb), non-WC-WC hydrogen bonds (_3D.pdb), energy of the Coulomb interactions (_Coulomb.pdb), van der Waals interactions (_VDW.pdb) and their sum (_stacking_sum.pdb). These files may be used for 3D visualizations, e.g., in VMD or Chimera.
.png files - pictures with 2D visualizations of computed parameters from Varna. Naming convention is similiar to the .pdb files.
_description - the main file containing the complete description of the structure. The file content starts from the list of all of the used parameters, followed by the list of helices, motifs, triplets and pseudo-knots. One can also find the list of both Watson-Crick (WC-WC) and non-Watson-Crick interactions, with the exact parameters of the hydrogen bonds. Additionally there is a simple dot-bracket representation of the secondary structure that can be used for visualization or energy computation by other tools ( _description example ).
_helices.csv - a file that contains a list of the deceted helices ( _helices.csv example).
_ion_pi.csv - a file that contains a list of the recognized ion-pi interactions (i.e., detected if a phosphorus atom in one nucleotide is placed close to the nucleobase of the second nucleotide) with their interaction energy values ( _ion_pi.csv example).
_motifs.csv - a file that contains list of the all RNA structural motifs ( _motifs.csv example)
_nucleotides_eval.csv - a file that contains the number of the WC-WC hydrogen bonds, non-WC-WC hydrogen bonds, Coulomb energy, van der Waals energy and their sum per nucleotide. The content gives the energetic description of the state of every nucleotide ( _nucleotides_eval.csv example).
_pairs.csv - a file that contains list of all detected pairs along with its classification according to edge (WC, Hoosteen, Sugar) and configuration (Cis or Trans) ( _pairs.csv example).
_pseudo.csv - a file that contains list of the all detected pseudo-knot ( _pseudo.csv example).
_stacking.csv - a file that contains a list of all recognized stacking pairs with their interaction energy values ( _stacking.csv example).
_triplets.csv - a file that contains the number of the recognized triplets ( _triplets.csv example).
_MINT.xls - a multiple sheets file that combines the above .csv files ( _MINT.xls example).
.pdb files - the analyzed structure with the column containing the beta-factors replaced with: the number of WC-WC hydrogen bonds (_2D.pdb), non-WC-WC hydrogen bonds (_3D.pdb), energy of the Coulomb interactions (_Coulomb.pdb), van der Waals interactions (_VDW.pdb) and their sum (_stacking_sum.pdb). These files may be used for 3D visualizations, e.g., in VMD or Chimera.
.png files - pictures with 2D visualizations of computed parameters from Varna. Naming convention is similiar to the .pdb files.
